Spectral Clustering of Protein Sequences
What is SCPS?
SCPS is an efficient, user-friendly, scalable and multi-platform implementation of a spectral clustering method for clustering homologous proteins. SCPS also implements connected component analysis and hierarchical clustering, integrates TribeMCL and interfaces with external tools such as Cytoscape and NCBI BLAST.
Overview
Clustering protein sequences based on their evolutionary relationship is important for sequence annotation as structural and functional relationships can potentially be inferred. Most of the existing methods are based on simply thresholding a measure related to the distance between sequences. Paccanaro et al (2006) mapped this problem into that of clustering the nodes of a weighted undirected graph in which each node corresponds to a protein sequence and the weights on the edges correspond to a measure of distance between two sequences. The goal is to partition such a graph into a set of discrete clusters whose members are homologs.
SCPS is an improved implementation of the method of Paccanaro et al (2006). The algorithm was tested on difficult sets of proteins whose relationships are known from the SCOP database. The method correctly identified many of the superfamily relationships, and the quality of the clusters as quantified by a measure that combines sensitivity and specificity was consistently better (on average, improvements were 45% over connected component analysis and 28% over TribeMCL).
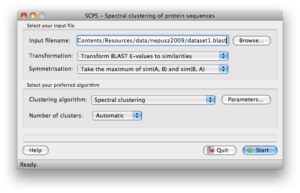
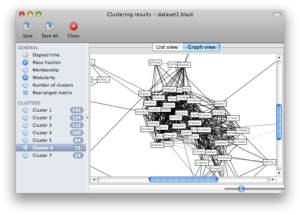
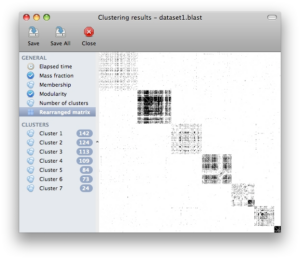
Screenshots
Click on any of the images below to get an idea of how SCPS looks like and whether it is suitable for you.
References
T. Nepusz, R. Sasidharan, and A. Paccanaro
SCPS: a fast implementation of a spectral method for detecting protein families on a genome-wide scale
BMC Bioinformatics, vol. 11, iss. 1, p. 120, 2010.![]()
![]()
Download
Windows installer (0.9.8)
Linux i386 archive (0.9.8)
Linux amd64 archive (0.9.8)
Mac OS X disk image (0.9.5)
Documentation
SCPS manual
Command line interface
Get the source code
SCPS is open-source. If you are interested in developing it further, feel free to download the source code and experiment with it. Patches, bug fixes are also appreciated. You will need Qt, FLENS, LAPACK, ARPACK and ARPACK++ and CMake to compile it.
Source code (0.9.8)
Bug reports, feedback
Something’s not working for you? Do you think you found an error? Do you want to contribute to the development of SCPS? Contact us!
![]()