Methods for denoising large scale protein-protein interaction experiments
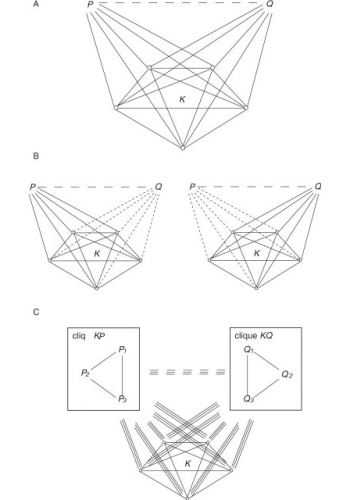
Large-scale experiments for the identification of protein-protein interactions are known to be error-prone. We have developed two methods for inferring protein-protein interactions which are based purely on topological properties of interaction networks observed experimentally and can correct many of the errors present in large-scale experiments. The basic idea of the first method (joint work with Haiyuan Yu, Harvard University and Valery Trifonov, Yale University) derives from the way in which large-scale experiments are carried out and particularly from the matrix model interpretation of their results. In these experiments, one protein (the bait), is used to pull out the set of proteins interacting with it (the preys), i.e. its protein complex, in the form of a list. When such lists differ only in a few elements, it is reasonable to assume that this is because of experimental errors, and the missing elements should therefore be added. Each list can be represented as a fully connected graph in which proteins occupy the nodes. Then the problem of identifying lists that differ in only a few elements is equivalent to finding a clique with a few missing edges, which we called a “defective clique”. Therefore the algorithm searches the network for defective cliques (i.e. nearly complete complexes of pairwise interacting proteins) and predicts the interactions that complete them. The second method (which we are currently refining), computes a diffusion distance between each pair of proteins and then infers an interaction when such distance is below a given threshold. Both methods have been shown to have a very good predictive performance, thus allowing the correction of many errors present in large-scale experiments.
H. Yu, A. Paccanaro, V. Trifonov, M. Gerstein (2006). Predicting interactions in protein networks by completing defective cliques. Bioinformatics 2006 Apr 1;22(7):823-9
A. Paccanaro, V. Trifonov, H. Yu, M. Gerstein (2005). Inferring protein-protein interactions using interaction network topologies — IJCNN 2005 – International Joint Conference on Neural Networks, Montreal, Canada