Prediction of gene essentiality from genomic features
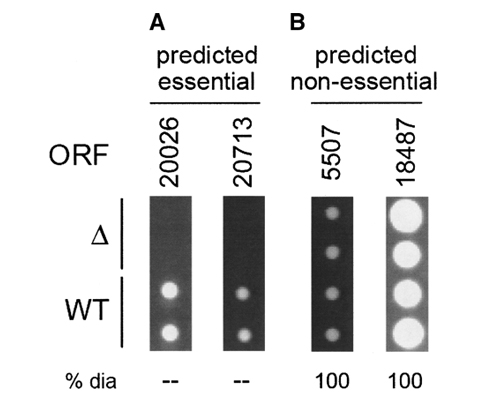
Predicting essential genes remains a main goal in directed drug design for antimicrobial or antifungal targets. Currently, most essential gene prediction is performed by homology searches to organisms where essentiality is known and has been experimentally tested. While quick and easy, this system is also simplistic. Together with Michael Seringhaus (Mark Gerstein’s Lab, Yale University), we aimed at improving the efficacy of such prediction through the integration of genomic-scale data, and the application of machine learning techniques. We trained a classification system on S. cerevisiae, where the Saccharomyces Genome Deletion project has ascertained essentiality for 95%+ of the genome. For each gene in the organism, we collected a set of genomic features – some derived from sequence information, others from functional genomics experiments. We used these data to learn a system that can predict essential genes in S. cerevisiae.
We then applied this system to three recently-sequenced yeast genomes (S. bayanus, S. mikatae, and C. albicans) for which essential genes have not been experimentally identified. We then compared our predictive engine to a simple BLAST homology search, and a subset of our putative essential candidates in S. bayanus and S. mikatae were tested with knockouts in vivo. We were able to demonstrate for the first time that it is possible to learn traits associated with essential genes in yeast species and to use these features in a predictive manner. Our approach therefore shows promise for the identification of drug targets in novel and pathogenic species. We are currently continuing this work by studying the relative importance and effects of the different types of features on the prediction. (This work was done in collaboration with the laboratories of Michael Snyder and Mark Gerstein, Yale University, USA).