 Department of
Computer Science
Department of
Computer Science
Code for Semantic Similarity Measures
This software returns semantics similarity scores for a
given gene list based on GO ontology files, and GO
Annotations files. There are 6 methods available in this
code: Resnik, Jiang, Lin, ISM_Resnik, ISM_Jiang, and
ISM_Lin, where ISM is the proposed method in the paper:
H. Yang, T. Nepusz, and A. Paccanaro, Improving GO semantic similarity measures by exploring the ontology beneath the terms and modelling uncertainty Bioinformatics, vol. 28, iss. 10, pp. 1383-1389, 2012.
The authors give permission for use, modification, and distribution of this software for any purpose.
In the default setting, the software uses the pre-downloaded GO ontology file, and the GO Annotations file. However, it provides an option of automatically downloading the recent GO ontology file. This software also allows the users to choose organisms, semantic similarity measures, and evidence codes ignored.
There are four main functions in this software: DOALL, INITIALIZE, GoSim, and GeneSim:
- INITIALIZE -- for a given, tree, it does all the steps necessary for semantic similarity calculation: parse the go ontology file, parse the go annotation file, get the number of genes annotated to a term, find the minimum number of annotations appearing in the least common ancester of two terms, and pre-compute all term similarities.
- DOALL -- it does all these things as INITIALIZE for three trees; moreover, it pre-computes gene product similarities.
- GoSim -- it returns the GO term similarity for the GoList.
- GeneSim -- it returns the gene product similarity for the GeneList
For a starter who wants only basic usage, see the examples below; for an advanced user, go to here to check the usage of these functions.
As this distribution contains some downloaded annotation files, and a downloaded GO ontology files, we acknowledge the Gene Ontology Consortium as the source of this software.
Download
We provide five sets of files for you to download:
- F1: contains the codes, some downloaded annotation files, and a downloaded go ontology file [download]
- F2: contains all files in F1, and pre-computed results for methods: Resnik, Jiang, Lin, ISM_Resnik, ISM_Jiang, and ISM_Lin [download]
- F3: contains files used for evaluation on sequence similarity [download]
- F4: contains files used for evaluation on PPI [download]
- F5: contains files used for evaluation on cell cycle [download]
Directory Structure
Codes and intermediate results are organised as follows:
- All code is stored inthe directory 'CODE_Version4'
- The default go ontology file, and the intermediate results related to go trees are stored in the directory 'GO_TREE'.
- For an organism, the default go annotation file, and the parsed results are stored in the directory 'GENE_OrganismName', e.g. 'GENE_Yeast' for Yeast (Saccharomyces cerevisiae)
- For an organism, the related intermediate results are stored in the directory 'GO2OrganismName', e.g. 'GO2Yeast' for Yeast (Saccharomyces cerevisiae)
Some examples to run the codes
How to use F1
To use the function GeneSim and GoSim, you need to run INITIALIZE or DOALL first. DOALL is specially suited for users who want to use the whole genomic similarity for all the trees because it pre-computes all term similarities and gene product similarities for all genes in the three trees. However, if you are only interested in a very small gene set for a given tree, use INITIALIZE instead. Some examples are listed below.
a. Extract files to the decretory 'GO_SIM_V4'
b. In Matlab environment, enter the directory 'GO_SIM_V4/CODE_Version4'
c. There are 6 methods available in this code. They are coded as 6 numbers: 1,2,3,18,28, and 38 representing Resnik, Jiang, Lin, ISM_Resnik, ISM_Jiang, and ISM_Lin respectively. The three tree 'biological_process', 'cellular_component', and 'molecular_function' are coded as 1, 2, and 3. If you want results for the organism 'Yeast' for Resnik's method, run
Which_Tree=1; INITIALIZE(1,-1,1, -1, 1,1, 'Yeast', [1],
-1,Which_Tree)
or
DOALL(1,-1,1, -1, 1,1, 'Yeast', [1], -1)
If you are curious about the parameter set, read the whole instruction or type 'help INITIALIZE' or 'help DOALL'
d. Then you can call function GeneSim to get the gene product similarity for a given gene list based on Resnik's method. For example, you have a gene list {'AAA1','AAC1','AAC2','AAC3'} and you are still in the directory 'GO_SIM_V4/CODE_Version4', and then run
SIM1 = GeneSim('..', {'AAA1','AAC1','AAC2','AAC3'}, 'biological_process', 1,'Yeast',-1)
You will get a similarity matrix on the gene list for biological_process based on Resnik's method
7.4979
1.5213 1.5788
2.2474
1.5213
8.3089 8.3089
8.3089
1.5788
8.3089 9.0020
9.0020
2.2474
8.3089 9.0020 9.0020
whose (i,j)-entry represents the semantic similarity between your i-th gene and j-th gene. The first parameter is the location of your GO_SIM_V4; and in this example, assuming that you are still in the directory 'CODE_Version4', it is '..'
Run
GoList={'GO:0000001','GO:0000002','GO:0000003','GO:0000011','GO:0000019','GO:0000022'};
SIMILARITY = GoSim('..', GoList, 1, 1,'Yeast',-1)
You will get a similarity matrix on the GO term list for biological_process based on Resnik's method
6.3279 4.1618
1.1590 5.4757
1.1590 2.6670
4.1618 6.0316
1.1590 2.6670
1.1590 2.6670
1.1590 1.1590
3.7393 1.1590
1.1590 1.1590
5.4757 2.6670
1.1590 6.8048
1.1590 2.6670
1.1590 1.1590
1.1590 1.1590
7.7493 1.1590
2.6670 2.6670
1.1590 2.6670
1.1590 6.5597
Congratulation! Now you become a basic user of this code. Up to now, you can obtain a similarity matrix based on Resnik for any give list in 'Yeast' for BP tree using the default setting for evidence codes and using the current ontology file and the annotation file.
Read the whole instruction to become an advanced user. Next you can exercise more.
e. If you want to a similarity matrix for 'cellular_component' and 'molecular_function' for the same gene list based on Resnik's method, run
SIM2 = GeneSim('..', {'AAA1','AAC1','AAC2','AAC3'},
'cellular_component', 1,'Yeast',-1)
%if you have run DOALL in step c; otherwise, you need to
first run
%Which_Tree=2; INITIALIZE(0,0,0, 0, 1,1, 'Yeast', [1],
-1,Which_Tree)
SIM3 = GeneSim('..', {'AAA1','AAC1','AAC2','AAC3'},
'molecular_function', 1,'Yeast',-1)
%if you have run DOALL in step c; otherwise, you need to
first run
%Which_Tree=3; INITIALIZE(0,0,0, 0, 1,1, 'Yeast', [1],
-1,Which_Tree)
f. If you want to do similar thing as above, but you want to try the ISM_Resnik (code: 18), then run
Which_Tree=1; INITIALIZE(0,0,0,0, 0,0, 'Yeast', [18],
-1,Which_Tree) or DOALL(0,0,0, 0, 0,0, 'Yeast',
[18], -1)
SIM1 = GeneSim('..', {'AAA1','AAC1','AAC2','AAC3'},
'biological_process', 18,'Yeast',-1)
Note that, assuming you have run step c, where some basic processes have been done for 'Yeast', you do not need to repeat them. This is why the first six parameters are set to 0. If you have not run step c before, then you need to run DOALL(1,-1,1, -1, 1,1, 'Yeast', [18], -1) instead of DOALL(0,0,0, 0, 0,0, 'Yeast', [18], -1); or INITIALIZE(1,-1,1, -1, 1,1, 'Yeast', [18], -1,Which_Tree) instead of INITIALIZE(0,0,0,0, 0,0, 'Yeast', [18], -1,Which_Tree).
g. If you want to use the code further for other organisms such as Arabidopsis (Arabidopsis thaliana), Mouse (Mus musculus), Worm (Caenorhabditis elegans) and Fly (Drosophila melanogaster), then do the similar things in steps c-d. For example, for 'Arabidopsis' and for Resnik's method, run
Which_Tree=1;INITIALIZE(0,0,1, -1, 1,1, 'Arabidopsis', [1],
-1,Which_Tree) or DOALL(0,0,1, -1, 1,1, 'Arabidopsis', [1],
-1)
SIM1 = GeneSim('..', yourgenelist, 'biological_process',
1,'Yeast',-1)
Note that, assuming you have run step c, where some basic processes have been done for the GO Ontology, you do not need to repeat them. This is why the first two parameters are set to 0. If you have not run step c before, then you need to run DOALL(1,-1,1, -1, 1,1, 'Arabidopsis', [1], -1).
h. For other organisms not in the list {Arabdopsis,
Yeast, Mouse, Worm, Fly}, we did not provide you an
annotation file, and you need to download the annotation
file by yourself. For example, if you downloaded the
annotation file for Ecoli (Escherichia coli) and
extracted it as 'your_annotation_file', then you can run
Which_Tree=1;INITIALIZE(0,0,1, 'your_annotation_file', 1,1,
'ECOLI', [1], -1,Which_Tree) or
DOALL(0,0,1,'your_annotation_file', 1,1, 'ECOLI', [1],
-1)
SIM1 = GeneSim('..', yourgenelist, 'biological_process',
1,'ECOLI',-1)
i. If you want to use the recent GO Ontology file to update the system, then the code can help download the recent GO Ontology file. Take 'Yeast' as an example, run
Which_Tree=1;INITIALIZE(1,0,1, -1, 1,1, 'Yeast', [1], -1,Which_Tree) or DOALL(1,0,1, -1, 1,1, 'Yeast', [1], -1)
Then, the code will download the recent GO Ontology file, and initialize the system.
j. If you have run (step c) already, and you do not want the default setting for evidence codes, then you can set your own evidence codes .For example, you want to ignore evidence codes IEA and NR, then you can run
Which_Tree=1;INITIALIZE(0,0,0, 0, 1,1, 'Yeast', [18], {'IEA', 'NR'},Which_Tree) or DOALL(0,0,0, 0, 1,1, 'Yeast', [18], {'IEA', 'NR'})
which means you do not parse the obo file and the annotation file again (because you did it in step c), and you ignore the annotations with evidence codes 'IEA' and 'NR', and you only want the result for the ISM_Resnik (18).
If you did not run (c) before, you want the same thing, then simply run
Which_Tree=1;INITIALIZE(1,-1,1, -1, 1,1, 'Yeast', [18], {'IEA', 'NR'}) or DOALL(1,-1,1, -1, 1,1, 'Yeast', [18], {'IEA', 'NR'})
Congratulation! Now you become an intermediate user of this code. Up to now, you can obtain a similarity matrix based on any of the available methods for any given gene list in any organism for any tree using any setting for evidence codes and using any version of the ontology file and any version of the annotation file.
How to use F2
All things are same as F1 except that it includes the intermediate results obtained by running
DOALL(1,-1,1, -1, 1,1, 'Yeast', [1,2,3,18,28,38], -1) ,
So you do not need the step c and f in F1.
How to use F3
0. Assume that you have downloaded F1, and initialized by running
DOALL(1,-1,1, -1, 1,1, 'Yeast', [1,2,3,18,28,38], -1) ,
Or you have download F2.
1. Extract files to the directory 'Yeast_Similarity'
2. Entry the directory 'Yeast_Similarity', run
exp_yeast('../GO_SIM_V4',0)
To get the correlation between sequence similarity and
semantic similarity WITHOUT the averaging procedure, where
'../GO_SIM_V4' is the path where the directory 'GO_SIM_V4'
is located. Here we assume that it is located in the parent
directory. In your machine, please specify it according to
where you put the GO_SIM_V4 code
3. Run
exp_yeast('../GO_SIM_V4',100)
To get the correlation between sequence similarity and semantic similarity WITH the averaging procedure, where 100 small intervals are created, and the means in these intervals are used to calculate the correlation.
How to use F4
0. Assume that you have downloaded F1, and initialized by running
DOALL(1,-1,1, -1, 1,1, 'Yeast', [1,2,3,18,28,38], -1) ,
Or you have download F2.
1. Extract files to the directory 'PPI'
2. Enter the directory 'PPI', run
for(method=[1,2,3,18,28,38]),
experiments('../GO_SIM_V4',1,method,-1), end
for(method=[1,2,3,18,28,38]),
experiments('../GO_SIM_V4',2,method,-1), end
for(method=[1,2,3,18,28,38]),
experiments('../GO_SIM_V4',3,method,-1), end
where '../GO_SIM_V4' is the path where the directory
'GO_SIM_V4' is located. Here we assume that it is located
in the parent directory. In your machine, please specify it
according to where you put the GO_SIM_V4 code
3. Run
makepic(1, 100)
To generate the ROC curve for BP tree, in which the number of points in the curve is less than to equal to 100. Run
makepic(2, 100)
To get the ROC curve for CC tree, and
makepic(3, 100)
To get the ROC curve for MF tree.
4. Run
cal_auc(0.002)
To get the AUC for BP, CC, and MF
How to use F5
0. Assume that you have downloaded F1, and initialized by running
DOALL(1,-1,1, -1, 1,1, 'Yeast', [1,2,3,18,28,38], -1) ,
Or you have download F2.
1. Extract files to the directory Cell_Cylce.
2. Entry the directory Cell_Cylce, run
exp_cellcycle('../GO_SIM_V4',0)
to get the correlation between microarray correlation and
semantic similarity WITHOUT the averaging procedure, where
'../GO_SIM_V4' is the path where the directory 'GO_SIM_V4'
is located. Here we assume that it is located in the parent
directory. In your machine, please specify it according to
where you put the GO_SIM_V4 code
3. Run
exp_cellcycle('../GO_SIM_V4',100)
to get the correlation between microarray correlation and
semantic similarity WITH the averaging procedure, where 100
small intervals are created, and the means in these
intervals are used to calculate the correlation.
Detailed Description of Some Functions
This page explains the functions of INITIALIZE, DOALL, GoSim, and GeneSim. To use the function GeneSim and GoSim, you need to down the data, and run INITIALIZE or DOALL first. In general, you need three steps:
Download the ontology file and the annotation file
You can skip this step, unless
a. you want to the recent ontology file, download the
file in OBO v1.2 format, from
http://www.geneontology.org/GO.downloads.ontology.shtml
b. you want to run the code for an organism not in the
list {Arabdopsis, Yeast, Mouse, Worm, Fly}, or
you want the recent annotation file for an organism in
the list {Arabdopsis, Yeast, Mouse, Worm, Fly},
you need to download the annotations file from
http://www.geneontology.org/GO.current.annotations.shtml
Call Function INITIALIZE
For a given, tree, INITIALIZE does all the steps
necessary for semantic similarity calculation: parse the
go ontology file, parse the go annotation file, get the
number of genes annotated to a term, find the minimum
number of annotations appearing in the least common
ancester of two terms, and pre-compute all term
similarities.
After calling INITIALIZE, you can use the function GoSim
to calculate the gene product similarities for a given
gene list, or use the function GoSim to retrieve the GO
term similarities for a given GO term list.
After calling INITIALIZE, some intermediate results are
stored. The advanced users may use these intermediate
results. The mat files for these results are described
below (the variables are shown in brackets):
OUT1: GOTable.mat (GOTable ColumName)
OUT2: Tree1.mat (Up, Down, GOList),Tree2.mat (Up, Down,
GOList),Tree3.mat (Up, Down, GOList).
GOList: GO id involved in the GO tree
Up{i}: is the index of i's parents in GOList
Down{i}: is the index of i's children in GOList
OUT3: ALLGOLIST.mat (ALLGOLIST)
ALLGOLIST(i,1): GO id including alternative GO id and
obsolete GO id
ALLGOLIST(i,2): which tree that ALLGOLIST(i,1) is
involved in; 0: none
ALLGOLIST(i,3): which position that ALLGOLIST(i,1) is
involved in tree
ALLGOLIST(i,2)
OUT4: AnnotationTable.mat (AnnotationTable)
AnnotationTable(i,:) contains information for annotation
i
OUT5: ALL_GENELIST_AltName.mat (ALL_GENELIST_AltName)
ALL_GENELIST_AltName(i,:) is the list of all alt name
of
ALL_GENELIST_AltName(i,1)
OUT6: GENE2GO.mat (GENE2GO)
GENE2GO{i,1} is the go term index list,
GENE2GO{i,2} is Evidence Code
OUT7: GENE2GO.mat (GENE2GO)
GO2GENE{i,1} is the gene index list in
ALL_GENELIST_AltName
GO2GENE{i,2} is Evidence Code
OUT8: LCA[123].mat ('TREE_LCA','SubGoList')
TREE_LCA(i,j): the minimum number of annotations
appearing in the least common ancester of SubGoList{i}
and SubGoList{j}
OUT9: GOSIM[1|2|3|18|28|38].mat ('SubGoList',
'GOSIM');
GOSIM(i,j) is the semantic similarity of SubGoList{i} and
SubGoList{j}
Usage
INITIALIZE(INIT_GO,GO_FILE,INIT_ANNO, ANNO_FILE,
INIT_TREE_P,INIT_TREE_LCA, Which_Organism, MethodList,
IgList)
if INIT_GO == 1, then the system will initialize
the go ontology using the go ontology file specified by
GO_FILE
if GO_FILE == -1, then use the default go ontology file
'../GO_TREE/IN/current.obo';;
if GO_FILE == 0, then download the obo file from
url='http://www.geneontology.org/ontology/obo_format_1_2/gene_ontology_ext.obo',
and store it as '../GO_TREE/IN/current.obo';
otherwise, use the gene_ontology file specified
INIT_ANNO == 1, then the system will initialize the go
annotation using the go annotation file specified by
ANNO_FILE
if ANNO_FILE==-1, then use the default annotation
file
Which_Organism=1 or 'Arabidopsis': gene_association.tair
(Arabidopsis thaliana)
Which_Organism=2 or 'Yeast': gene_association.sgd
(Saccharomyces cerevisiae)
Which_Organism=3 or 'Mouse': gene_association.mgi
(Mus musculus)
Which_Organism=4 or 'Worm': gene_association.wb
(Caenorhabditis elegans)
Which_Organism=5 or 'Fly': gene_association.fb
(Drosophila melanogaster)
ANNO_FILE is the annotation file specified,
otherwise.
if INIT_TREE_P ==1, then the number of genes annotated to
each go term is calculated
if TREE_LCA ==1, then the least common ancester is
calculated
Which_Organism is either a number in [1,2,3,4,5] or an
organism name in the list
{'Arabidopsis','Yeast','Mouse','Worm','Fly'} or any
specified organism name.
Which_Tree=1: 'biological_process'
Which_Tree=2:'cellular_component'
Which_Tree=3:'molecular_function'
MethodList: a subset of
[1,2,3,18,28,38,19,29,39,15,25,35]
1: Resnik (1995)
2: Jiang and Conrath (1997) alpha=0; beta=1; T=1;
3: Lin (1998)
18: ISM_Resnik
28: ISM_Jiang
38: ISM_Lin
use the default ignore list={'IEA'; 'NR'; 'ND'; 'IC'}, if
IgList==-1;
Use the ignore list specified by IgList, otherwise.
Example:
Which_Tree=1;
INITIALIZE(1,-1,1, -1, 1,1, 'Yeast',
[1,2,3,18,28,38], -1,Which_Tree)
will do the following for Yeast for
'biological_process'
a1: initialize the go ontology using the default
file ../GO_TREE/IN/current.obo
a2: initialize the go annotation using the default
go annotation file
../GENE_Yeast/IN/gene_association.sgd
a3: the number of genes annotated to each go term
is calculated
a4: the least common ancester is calculated
a5: calculate the GO terms semantic similarity
using methods 1,2,3,18,28,and 38 ignoring the evidence
codes IEA, NR, ND, and IC
Call Function DOALL
DOALL does all these things as INITIALIZE for three trees; moreover, it pre-computes gene product similarities. Therefore there are more outputs:
OUT10: SIM[1|2|3|18|28|38].mat ('GENELIST', 'SIM');
SIM(i,j) is the semantic similarity of GENELIST{i} and
GENELIST{j}
Example:
DOALL(1,-1,1, -1, 1,1, 'Yeast', [1,2,3,18,28,38], -1)
will do the following for Yeast for all three trees:
a1: initialize the go ontology using the default file
../GO_TREE/IN/current.obo
a2: initialize the go annotation using the default go
annotation file ../GENE_Yeast/IN/gene_association.sgd
a3: the number of genes annotated to each go term is
calculated
a4: the least common ancester is calculated
a5: calculate the GO terms semantic similarity using
methods 1,2,3,18,28,and 38 ignoring the evidence codes
IEA, NR, ND, and IC
a6: calculate the gene products semantic similarity using
methods 1,2,3,18,28,and 38 ignoring the evidence codes
IEA, NR, ND, and IC
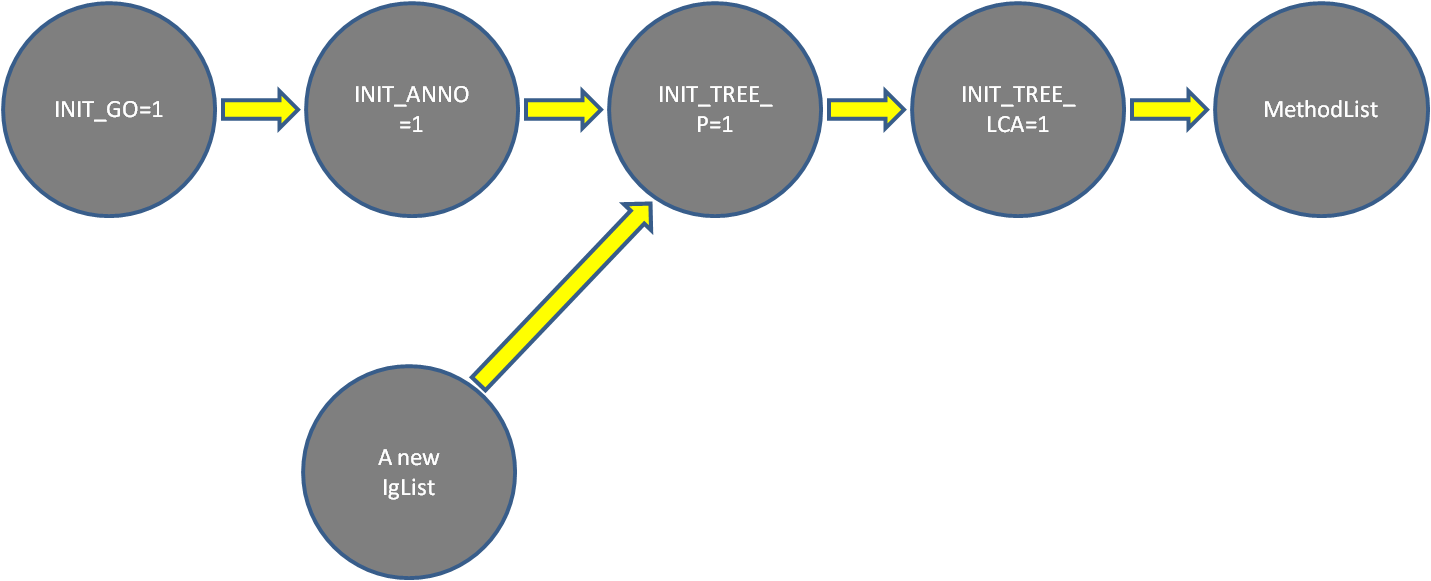
The picture below shows the dependency between the
parameters in the function INITIALIZE and DOALL: if
one parameter is set to 1 or reset, then parameters of
all its descendents except for MethodList should be set
1. For example, if INIT_GO == 1, then a
new GO ontology is introduced, and all others parts
should be re-structured, and should be set.
Another example, if you want to use a new Iglist {'IEA'},
then only INIT_TREE_P and INIT_TREE_LCA are affected, and
should be set to 1, and consequently, the results will be
updated. Because INIT_GO and INIT_ANNO are not affected
by a new setting of Iglist, they should be set 0 if you
run the code before.
Call Function GoSim
GoSim returns the GO term similarity for the GoList
Usage
!!!If you want to use the updated data, or you do
not download/have the intermediate results, or you want
to call this function for a new organism, initialize the
system using the function INITIALIZE first before calling
the function GoSim
SIMILARITY = GoSim(Root, GoList, Which_Tree,
Which_Method,Which_Organism,IgList)
SIMILARITY(i,j) is the semantic similarity of GoList{i}
and GoList{j}
Root is path of the code; if Root==-1, then use the
default Root='..', assuming that you are in the
directory 'CODE_Version4'
GoList is a cell array like
{'GO:0000001','GO:0000002','GO:0000003','GO:0000011','GO:0000019','GO:0000022'}
Which_Tree=1: 'biological_process'
Which_Tree=2:'cellular_component'
Which_Tree=3:'molecular_function'
Which_Method=1: Resnik (1995)
Which_Method=2: Jiang and Conrath (1997) alpha=0; beta=1;
T=1;
Which_Method=3: Lin (1998)
Which_Method=18: ISM_Resnik
Which_Method=28: ISM_Jiang
Which_Method=38: ISM_Lin
Which_Organism is either a number in [1,2,3,4,5] or the
organism name
Which_Organism=1: Arabidopsis (Arabidopsis
thaliana);
Which_Organism=2: Yeast (Saccharomyces
cerevisiae);
Which_Organism=3: Mouse (Mus musculus)
Which_Organism=4: Worm (Caenorhabditis
elegans)
Which_Organism=5: Fly (Drosophila
melanogaster)
use the default Ignore List={'IEA'; 'NR'; 'ND'; 'IC'}, if
IgList==-1
Use the specified by IgList otherwise
Example:
GoList={'GO:0000001','GO:0000002','GO:0000003','GO:0000011','GO:0000019','GO:0000022'};
SIMILARITY = GoSim('..', GoList, 1, 1,'Yeast',-1)
which means that you are in the directory
'CODE_Version4', and you want to get the similarity
scores for the GO term list
{'GO:0000001','GO:0000002','GO:0000003','GO:0000011','GO:0000019','GO:0000022'}
using Resnik's method for BP tree ignoring the
annotation with evidence codes {'IEA'; 'NR'; 'ND';
'IC'}
Call Function GeneSim
GeneSim returns the gene product similarity for the
GeneList
Usage
!!!If you want to use the updated data, or you do not
download/have the intermediate results, or you want to
call this function for a new organism, initialize the
system using the function INITIALIZE first before calling
the function GeneSim
SIMILARITY = GeneSim(Root, GeneList, Which_Tree,
Which_Method,Which_Organism,IgList)
SIMILARITY(i,j) is the semantic similarity of GeneList{i}
and GeneList{j}
Root is path of the code; if Root==-1, then use the
default Root='..', assuming that you are in the directory
'CODE_Version4'
GeneList is a cell array like
{'15S_RRNA','21S_RRNA','AAC1','AAC3','AAD10','AAD14','AAD15','AAD16'}
Which_Tree=1: 'biological_process'
Which_Tree=2:'cellular_component'
Which_Tree=3:'molecular_function'
Which_Method=1: Resnik (1995)
Which_Method=2: Jiang and Conrath (1997) alpha=0; beta=1;
T=1;
Which_Method=3: Lin (1998)
Which_Method=18: ISM_Resnik
Which_Method=28: ISM_Jiang
Which_Method=38: ISM_Lin
Which_Organism is either a number in [1,2,3,4,5] or the
organism name
Which_Organism=1: Arabidopsis (Arabidopsis
thaliana);
Which_Organism=2: Yeast (Saccharomyces
cerevisiae);
Which_Organism=3: Mouse (Mus musculus)
Which_Organism=4: Worm (Caenorhabditis
elegans)
Which_Organism=5: Fly (Drosophila
melanogaster)
use the default Ignore List={'IEA'; 'NR'; 'ND'; 'IC'}, if
IgList==-1
Use the specified by IgList otherwise
Example:
list={'15S_RRNA','21S_RRNA','AAC1','AAC3','AAD10','AAD14','AAD15','AAD16'}
SIMILARITY = GeneSim('..', list, 1, 1,'Yeast',-1)
which means that you are in the directory
'CODE_Version4', and you want to get the similarity
scores for the genelist {'AAA1','AAC1','AAC2','AAC3'}
using Resnik's method for BP tree ignoring the annotation
with evidence codes {'IEA'; 'NR'; 'ND'; 'IC'}

