Techniques for integrating different protein-protein interaction experiments
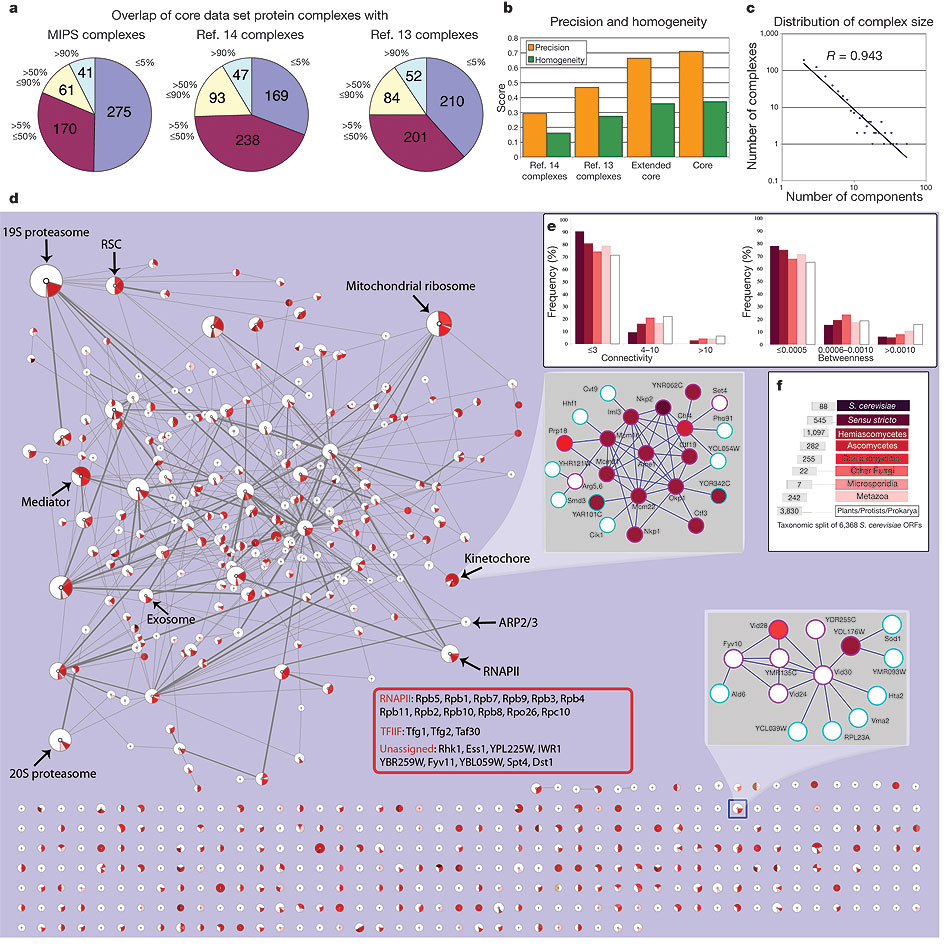
The ultimate goal of functional genomics is to discover the functions of all proteins in a genome. Proteins carry out their molecular functions by interacting with other molecules, mainly other proteins. Thus, protein interactions provide an important clue to the function of proteins. Many interaction datasets, mostly from large-scale experiments are available now. However, the quality of protein interaction experiments varies greatly with the researcher who performs the experiment and with the particular technique used. Therefore, it is important to be able to integrate the results of different experiments into a more reliable measurement. Together with Haiyuan Yu (Harvard University), we developed a system that provided such reliable measurement by combining the results of different experiments through a set of learned weights. Using the hand-curated protein complexes in the MIPS reference database we trained a system to assign a probability that each pairwise interaction is true based on experimental reproducibility and mass spectrometry scores from the relevant purifications.
Thus from the raw experimental data we obtained a protein–protein interaction network as an undirected weighted graph in which individual proteins are nodes and the weight of the edge connecting two nodes is the probability that the interaction is correct. We are currently attempting to improve these results by including information about the network topology. (This work is done in collaboration with the laboratory of Andrew Emili at the University of Toronto, Canada.)